The dilemma
Have you, or a cattle producer friend, ever encountered a situation where you vaccinated your cow herd against rotavirus scours but still had problems with rotavirus scours? What about experiencing a Histophilus somni (H. somni) break in pens of vaccinated feedlot calves? Or Mycoplasma bovis (M. bovis)? Or pinkeye?
There are several possible contributing factors that should be investigated in these types of cases, but each one of these disease-causing organisms, or pathogens, exhibits strain variation. This strain variation might explain why the vaccines used did not protect against the infecting organisms.
The importance of the 'right vaccine'
The appropriate vaccination program is a crucial component of an effective preventive health program. It is important to work closely with your veterinarian to select the correct vaccines and determine the proper timing of administration. Vaccine selection and correct use are essential to optimizing protection against infectious diseases.
One of the crucial principles of vaccination is the goal of stimulating immunity against the specific disease-causing organism, or pathogen, the animal may encounter. So it is crucial to use a vaccine that matches the specific pathogens. If we are not using vaccines designed to stimulate protection against the appropriate viruses and bacteria, we are wasting time and money.
We must be aware of the disease challenges facing our cattle: What viruses or species of bacteria are most likely to be a threat? Similarly, if we’ve had disease problems in the past in our cattle, what pathogens were diagnosed? These are situations where it is essential to work with your veterinarian to assess the clinical history, evaluate sick cattle, perform necropsies and submit samples to a diagnostic laboratory.
Vaccines contain antigens, usually made up of proteins, specific to a given virus or bacteria used to manufacture the product. When administered properly, the immune system will recognize the vaccine antigen and develop a response specific to it. The antibodies produced by the calf’s or cow’s body are specific to the antigen (or strain proteins) in the vaccine.
Figure 1 is a schematic representation of the antigens from several different strains of the same species of bacteria. Antibodies specific to one type of antigen for one strain will not match up with the antigen for another strain, even within the same species.

The specificity of that immune response is a critical aspect of the process and can be compared to a lock and key model. If you don’t have the right key, it won’t match up with the lock. Even though different locks may appear to be the same, there are slight variations that will only match up with the right key. This “lock and key” concept applies to vaccine antigens and the antibodies produced by the animal.
When we realize that immunity is strain-specific, with little or no cross-protection, we can understand that the vaccine must contain the appropriate antigens representing the desired strain to be effective. If an animal has been vaccinated against one strain but is challenged with a different strain of the same virus or bacterial species, the immune system may not recognize it and the animal may not be protected.
Strain variation is a consideration with many different cattle pathogens, including H. somni, M. bovis and rotavirus, as well as the bacteria that are associated with pinkeye. For example, strain variation associated with H. somni may explain why a calf may be infected by the bacteria if the vaccine strain was different from the strain that infected the calf. If a commercial, off-the-shelf vaccine isn’t effective, your veterinarian may recommend developing an autogenous or custom-made vaccine made from the appropriate strains.
Genes and strain variation
To determine the specific cause of an infectious disease syndrome in cattle, a veterinarian will assess the clinical signs and history, as well as perform a postmortem necropsy on any dead animals and/or take samples from live animals. Those samples are then submitted to a veterinary diagnostic laboratory.
Once the specific pathogen has been identified, additional testing can be performed using molecular biology to better characterize the organism, determining its “genetic fingerprint.” This process is called whole-genome sequencing. The genes of the pathogen determine what antigens it will possess, much as our genes determine if we are blonde or brunette, or if we are tall or short. The gene sequencing results allow us to characterize and compare the organisms with others, assessing the predicted antigenic or strain differences.
If we want a vaccine that will stimulate immunity against a specific strain of bacteria, we need to make sure we utilize that strain when we manufacture the product. Knowing the genetic makeup of the organism will enable us to determine if it will match up strain-wise and enable us to select the correct isolate to make the vaccine.
A good example of a pathogen that exhibits strain variation is Histophilus somni (H. somni), the bacteria associated with a variety of disease syndromes in cattle including respiratory disease, brain infections, infection of the myocardium (in the heart) and arthritis. We utilize whole-genome sequencing to evaluate H. somni isolates, to characterize them genetically and compare them for genetic differences associated with antigenic variation or strain differences.
The genetic information is presented in a dendrogram (or phylogenetic tree), similar to a family tree. This example (Figure 2) of a dendrogram shows multiple H. somni isolates from various feedlots. The different groups of isolates would represent clades or clusters or strains.
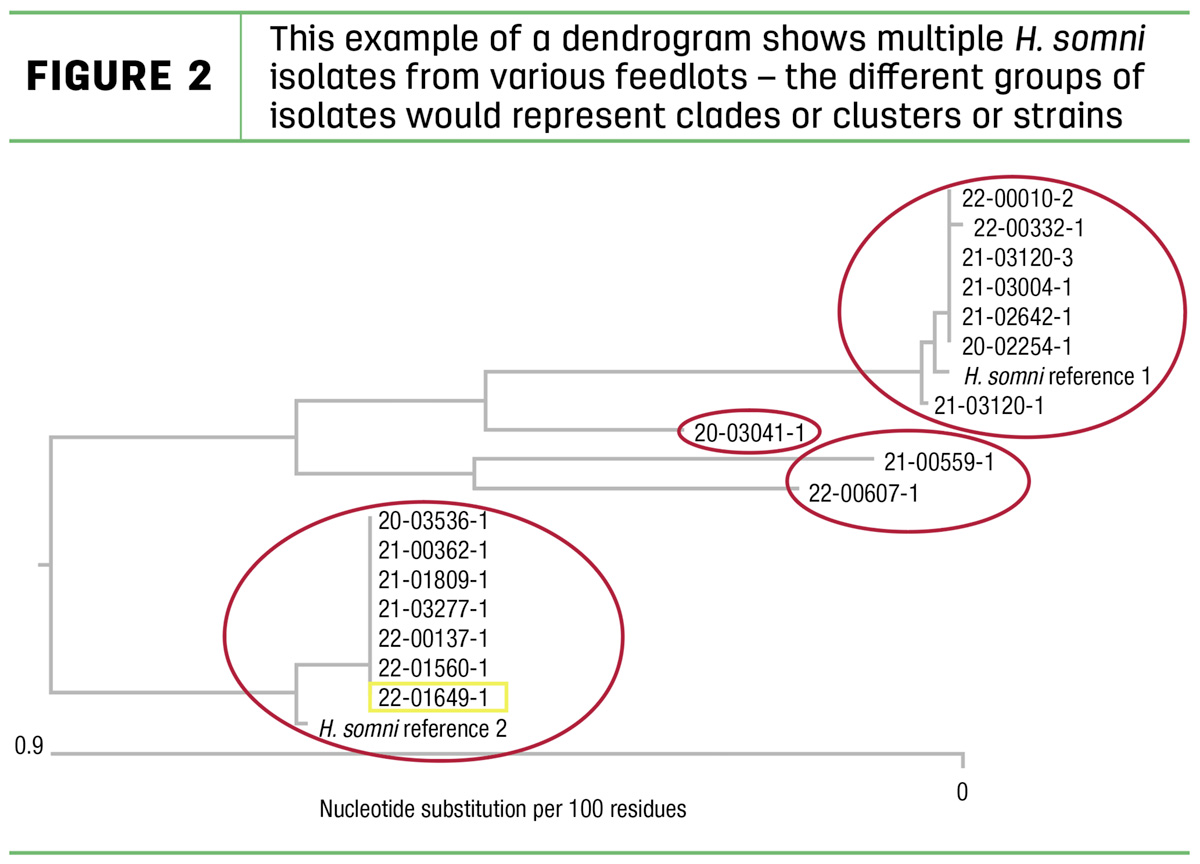
Each strain group, circled in red, includes isolates that are very similar genetically. Because of the similarity of these isolates, we can assume they are very similar antigenically and would be protected by the same antibody. So one isolate out of each strain group could be expected to stimulate immunity protective against all the isolates in that group.
However, isolates from different strain groups would be significantly different antigenically. An antibody specific to one strain group would not protect against an isolate from another strain group. Using our lock and key analogy, an isolate from one strain group would represent a different lock than an isolate from another strain group. So the same antibody “key” would not be expected to match up with two different strain “locks.”
The dendrogram is a tool for selection of isolates to manufacture a vaccine that will stimulate the desired specific immune response, covering the strain variation in the entire group of isolates. If we want to manufacture a vaccine for cattle to protect them against all four of these specific strains, we would select one isolate from each strain group to manufacture a combination product. Autogenous or custom-made vaccines are well suited to this approach, allowing us to vaccinate against the specific strains affecting the cattle.
Conclusion
To effectively stimulate the desired immune response when vaccinating cattle, it is important the vaccine contains the appropriate strains that are most likely to affect your cattle. Diagnostic lab work, including next-generation whole-genome sequencing, helps to determine what pathogens are involved and if strain variation should be considered.
Some pathogens exhibit considerable strain variation, and commercial “off-the-shelf” vaccines may not contain the necessary strains. An important option to consider and discuss with your veterinarian is using an autogenous vaccine with the appropriate specific isolates and strains. This allows you to develop a preventive health program customized to your cattle and your operation.










